8. Tips and Tricks
8.1. Getting Bad Pixel Masks from the archive
Starting with DRAGONS v3.1, the static bad pixel masks (BPMs) are now handled as calibrations. They are downloadable from the archive instead of being packaged with the software. There are various ways to get the BPMs.
Note that at this time there no static BPMs for Flamingos-2 data.
8.1.1. Manual search
Ideally, the BPMs will show up in the list of associated calibrations, the “Load Associated Calibration” tab on the archive search form (next section). This will happen of all new data. For old data, until we fix an issue recently discovered, they will not show up as associated calibration. But they are there and can easily be found.
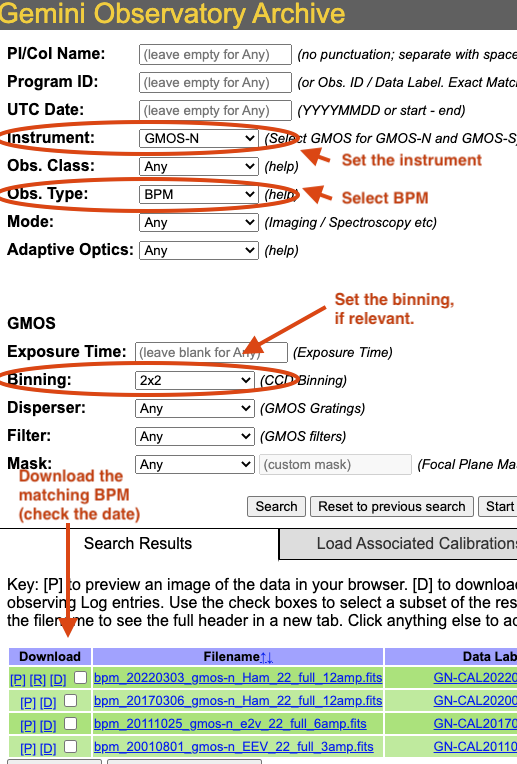
On the archive search form, set the “Instrument” to match your data, set the “Obs.Type” to “BPM”, if relevant for the instrument, set the “Binning”. Hit “Search” and the list of BPMs will show up as illustrated in the figure below.
The date in the BPM file name is a “Valid-from” date. It is valid for data taken on or after that date. Find the one most recent BPM that is valid for your date and download (click on “D”) it. Then follow the instructions found in the tutorial examples.
8.1.2. Associated calibrations
The BPMs are now handled like other calibrations. This means that they are also downloaded from the archive. From the archive search form, once you have identified your science data, select the “Load Associated Calibrations” (which turns to “View Calibrations” once the table is loaded). The BPM will show up with the green background.
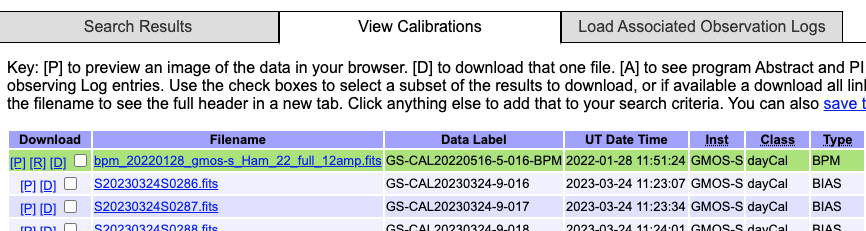
This will be the case for new data (from late March 2023). For old data, until we fix an issue recently discovered, they will not show up as associated calibration and you will have to find them manual as explained in the previous section, Manual search. We understand the issue and are working on a fix.
8.1.3. Calibration service
The calibration service in DRAGONS 3.1 adds several new features. One of them is the ability to search multiple databases in a serial way, including online database, like the Gemini archive.
The system will look first in your local database for processed calibration
and BPMs. If it does not find anything that matches, it will look in the
next database. To activate this feature, in ~/.dragons/, create or edit
the configuration file dragonsrc as follows:
[calibs]
databases = ${path_to_my_data}/niriimg_tutorial/playground/cal_manager.db get store
https://archive.gemini.edu get
If you know that you will be connected to the internet when you reduce the data, you do not need to pre-download the BPM, DRAGONS will find it for you in the archive.
If you want to pre-download the BPM without having to search for it, like in the previous two sections, you can let DRAGONS find it and download it for you:
$ reduce -r getBPM <file_for_which_you_need_bpm>
$ caldb add calibrations/processed_bpm/<the_bpm>
8.2. Plot a 1-D spectrum
Here is how to plot an extracted spectrum produced by DRAGONS with Python and matplotlib.
1import matplotlib.pyplot as plt
2import numpy as np
3
4import astrodata
5import gemini_instruments
6
7ad = astrodata.open('S20171022S0087_1D.fits')
8ad.info()
9
10data = ad[0].data
11wavelength = ad[0].wcs(np.arange(data.size)).astype(np.float32)
12units = ad[0].wcs.output_frame.unit[0]
13
14# add aperture number and location in the title.
15# check that plt.xlabel call. Not sure it's right, it works though.
16plt.xlabel(f'Wavelength ({units})')
17plt.ylabel(f'Signal ({ad[0].hdr["BUNIT"]})')
18plt.plot(wavelength, data)
19plt.show()
8.3. Inspect the sensitivity function
Plotting the sensitivity function is not obvious. Using Python, here’s a way to do it.
1from scipy.interpolate import BSpline
2import numpy as np
3import matplotlib.pyplot as plt
4
5import astrodata
6import gemini_instruments
7
8ad = astrodata.open('S20170826S0160_standard.fits')
9
10sensfunc = ad[0].SENSFUNC
11
12order = sensfunc.meta['header'].get('ORDER', 3)
13func = BSpline(sensfunc['knots'].data, sensfunc['coefficients'].data, order)
14std_wave_unit = sensfunc['knots'].unit
15std_flux_unit = sensfunc['coefficients'].unit
16
17w1 = ad[0].wcs(0)
18w2 = ad[0].wcs(ad[0].data.size)
19
20x = np.arange(w1, w2)
21plt.xlabel(f'Wavelength ({std_wave_unit})')
22plt.ylabel(f'{std_flux_unit}')
23plt.plot(x, func(x))
24plt.show()
In the science-approved version of the GMOS longslit support in DRAGONS, there will be an interactive tool to inspect and adjust the sensitivity function.