5. Tips and Tricks
This is a collection of tips and tricks that can be useful for reducing different data, or to do it slightly differently from what is presented in the example.
5.1. Getting Bad Pixel Masks from the archive
Starting with DRAGONS v3.1, the static bad pixel masks (BPMs) are now handled as calibrations. They are downloadable from the archive instead of being packaged with the software. There are various ways to get the BPMs.
Note that at this time there no static BPMs for Flamingos-2 data.
5.1.1. Manual search
Ideally, the BPMs will show up in the list of associated calibrations, the “Load Associated Calibration” tab on the archive search form (next section). This will happen of all new data. For old data, until we fix an issue recently discovered, they will not show up as associated calibration. But they are there and can easily be found.
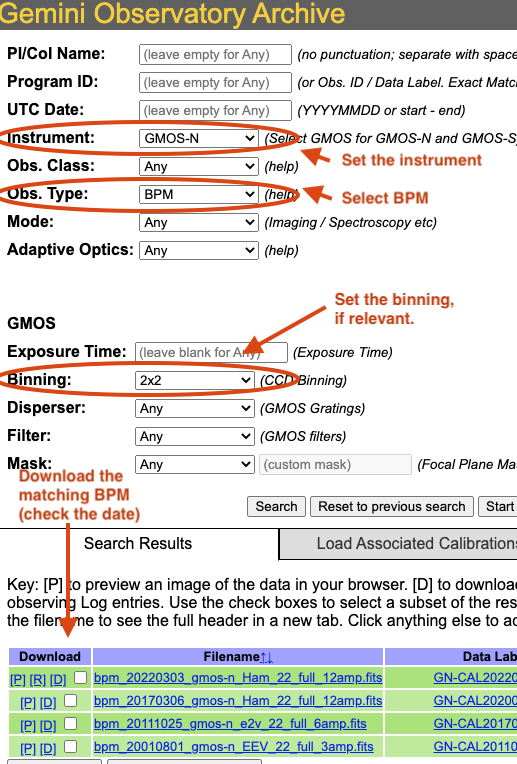
On the archive search form, set the “Instrument” to match your data, set the “Obs.Type” to “BPM”, if relevant for the instrument, set the “Binning”. Hit “Search” and the list of BPMs will show up as illustrated in the figure below.
The date in the BPM file name is a “Valid-from” date. It is valid for data taken on or after that date. Find the one most recent BPM that is valid for your date and download (click on “D”) it. Then follow the instructions found in the tutorial examples.
5.1.2. Associated calibrations
The BPMs are now handled like other calibrations. This means that they are also downloaded from the archive. From the archive search form, once you have identified your science data, select the “Load Associated Calibrations” (which turns to “View Calibrations” once the table is loaded). The BPM will show up with the green background.
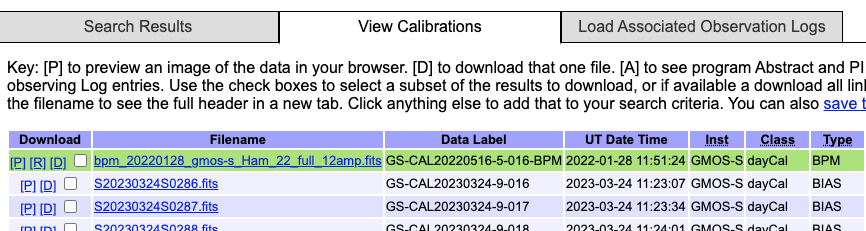
This will be the case for new data (from late March 2023). For old data, until we fix an issue recently discovered, they will not show up as associated calibration and you will have to find them manual as explained in the previous section, Manual search. We understand the issue and are working on a fix.
5.1.3. Calibration service
The calibration service in DRAGONS 3.1 adds several new features. One of them is the ability to search multiple databases in a serial way, including online database, like the Gemini archive.
The system will look first in your local database for processed calibration
and BPMs. If it does not find anything that matches, it will look in the
next database. To activate this feature, in ~/.dragons/, create or edit
the configuration file dragonsrc as follows:
[calibs]
databases = ${path_to_my_data}/niriimg_tutorial/playground/cal_manager.db get store
https://archive.gemini.edu get
If you know that you will be connected to the internet when you reduce the data, you do not need to pre-download the BPM, DRAGONS will find it for you in the archive.
If you want to pre-download the BPM without having to search for it, like in the previous two sections, you can let DRAGONS find it and download it for you:
$ reduce -r getBPM <file_for_which_you_need_bpm>
$ caldb add calibrations/processed_bpm/<the_bpm>
5.2. Create Master Fringe Frame
The reduction of some datasets requires a master fringe frame. The filters that need a fringe frame are shown in the appendix Fringe Correction Tables.
To create the master fringe frame from the dithered science observations and add it to the calibration database:
$ reduce @list_of_science.txt -r makeProcessedFringe
This command line will produce an image with the _fringe suffix in the
current working directory.
Again, note that this step is only needed for images obtained with some detector and filter combinations. Make sure you checked the Fringe Correction Tables.
The above can be done with the API as follows:
1reduce_fringe = Reduce()
2reduce_fringe.files.extend(list_of_science)
3reduce_fringe.recipename = 'makeProcessedFringe'
4reduce_fringe.runr()
5.3. Bypass automatic calibration association
We can think of two reasons why a user might want to bypass the calibration manager and the automatic processed calibration association. The first is to override the automatic selection, to force the use of a different processed calibration than what the system finds. The second is if there is a problem with the calibration manager and it is not working for some reason.
Whatever the specific situation, the following syntax can be used to bypass the calibration manager and set the input processed calibration yourself.
$ reduce @sci_images.list --user_cal processed_bias:S20001231S0001_bias.fits processed_flat:S20001231S0002_flat.fits
The list of recognized processed calibration is:
processed_arc
processed_bias
processed_dark
processed_flat
processed_fringe
processed_standard
5.4. Browse Recipes and Primitives
“reduce”, either the command line or the API class, is the tool that selects and run a “recipe”. A recipe is a sequence of operations called “primitives”. Each primitives has a defined set of input parameters with default values that can be overriden by the user.
The “showrecipes” command line is used to show the default recipe for a file, a specific recipe for that file, or all the recipes associated with the file.
Once you know the recipe and primitives it is calling, you can explore the primitives’ parameters using the “showpars” command line.
The tools are fully documented in the Recipe System Users Manual.
5.5. Customizing input parameters
From the command line, setting the value of a primitive input parameter is done as follows:
$ reduce @sci.lis -p stackFrames:scale=True
The -p flag indicates that the following items are parameter changes. The
syntax is <primitive_name>:<parameter_name>=<value>
From the API, the uparms attribute to the Reduce instance is used.
1reduce_science.uparms.append(("stackFrames:scale", True))
5.6. Setting the output suffix
When troubleshooting an issue or trying various settings to optimize a reduction, it might be useful to name the final recipe output differently for each attempt.
Only the suffix of the final output file can be changed, not its full name.
From the command line:
$ reduce @sci.lis --suffix='newsuffix'
From the API:
1reduce_science.suffix = "newsuffix"
2reduce_science.runr()